

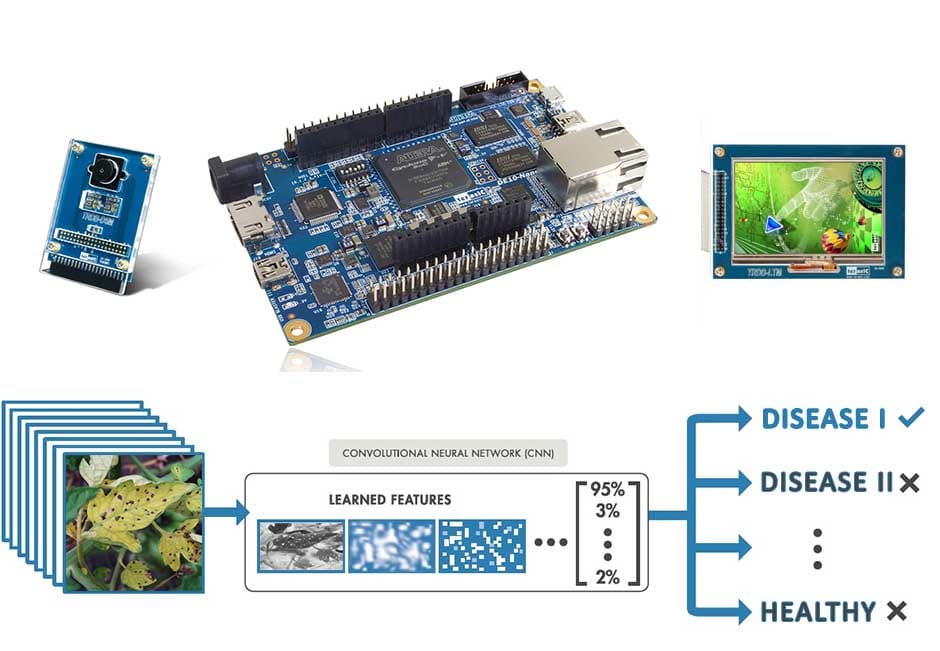
AP074 » Detection and Recognition of Plant Diseases using FPGA based real-time processing
Deep Learning dominates contemporary Machine Vision, with Convolutional Neural Networks (CNNs) being the state-of-the-art recognition system. In our work, we attempt to detect and accurately recognize diseases in plants, given a considerable dataset of images. Our focus would be on recognizing exact diseases by applying image processing techniques to close up images of plant leaves. We employ FPGA for real-time inference of the trained CNNs. For detection, we obtain multi-spectral images (visual/RGB) and calculate the NDVI index for an aerial video feed. Areas beyond a threshold are marked sick, and a map is generated with locations of affected regions.
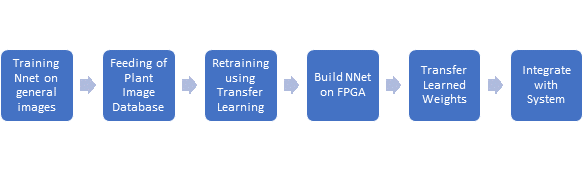
We treat the recognition of different diseases as a classification task, and train a CNN using a collected dataset. We would be training separate CNNs for each plant type, each of which would be able to recognize the diseases relevant to that plant species. Due to the limited quantity of training data and the nature of this task, we focus on using transfer learning for training the CNN, where we already provide it with knowledge regarding general images, and use plant disease specific data only for determining very high level features. In addition, we plan to look at active learning to find the most required images to be labelled, as more similar data can be collected, instead of blanket data collection. The inference using this trained Neural Network will be done based on FPGA as real-time performance is expected for this computationally heavy task.
Our system will be in the form of a single-board computer based device using FPGA for complex computations. Images obtained would be processed on the device itself, mostly using the FPGA. As inference in CNNs is done using FPGA, this is possible in real-time. We are testing Inception, RESNET and Alexnet CNN architectures and will carry out training of the Neural Network on a high performance device (one time task). The initial case would be done using python with tensorflow, opencv, and numpy. The trained neural network architecture would be rebuilt in low-level, and the trained-weights would be borrowed and directly used.
Our key beneficiaries include small-scale home gardeners (people without access to specialized knowledge regarding plant diseases), green-house based farmers, and plant disease research groups.
Possible future extensions for this work include detecting diseases (anomaly detection) using general aerial images. Considering the ubiquity of drone based agriculture for large fields, this will show emerge invaluable in near future.
Deep Learning dominates contemporary Machine Vision, with Convolutional Neural Networks (CNNs) being the state-of-the-art recognition system. In our work, we attempt to detect and accurately recognize diseases in plants, given a considerable dataset of images.

Our focus would be on recognizing exact diseases by applying image processing techniques to close up images of plant leaves. We employ FPGA for real-time inference of the trained CNNs as well as real time image processing. The performance boosts obtainable through the use of Altera FPGA devices would be sufficient to build this product as a feasible device.
For detection, we obtain multi-spectral images (visual/RGB) and calculate the NDVI index for an aerial video feed. Areas beyond a threshold are marked sick, and a map is generated with locations of affected regions. Part of the image processing would be carried out on FPGA to ensure real-time performance.
We treat the recognition of different diseases as a classification task, and train a CNN using a collected dataset. We would be training separate CNNs for each plant type, each of which would be able to recognize the diseases relevant to that plant species. Due to the limited quantity of training data and the nature of this task, we focus on using transfer learning for training the CNN, where we already provide it with knowledge regarding general images, and use plant disease specific data only for determining very high level features. In addition, we plan to look at active learning to find the most required images to be labelled, as more similar data can be collected, instead of blanket data collection. The inference using this trained Neural Network will be done based on FPGA as real-time performance is expected for this computationally heavy task.
Our system will be in the form of a single-board computer based device using FPGA for complex computations. Images obtained would be processed on the device itself, mostly using the FPGA. As inference in CNNs is done using FPGA, this is possible in real-time.
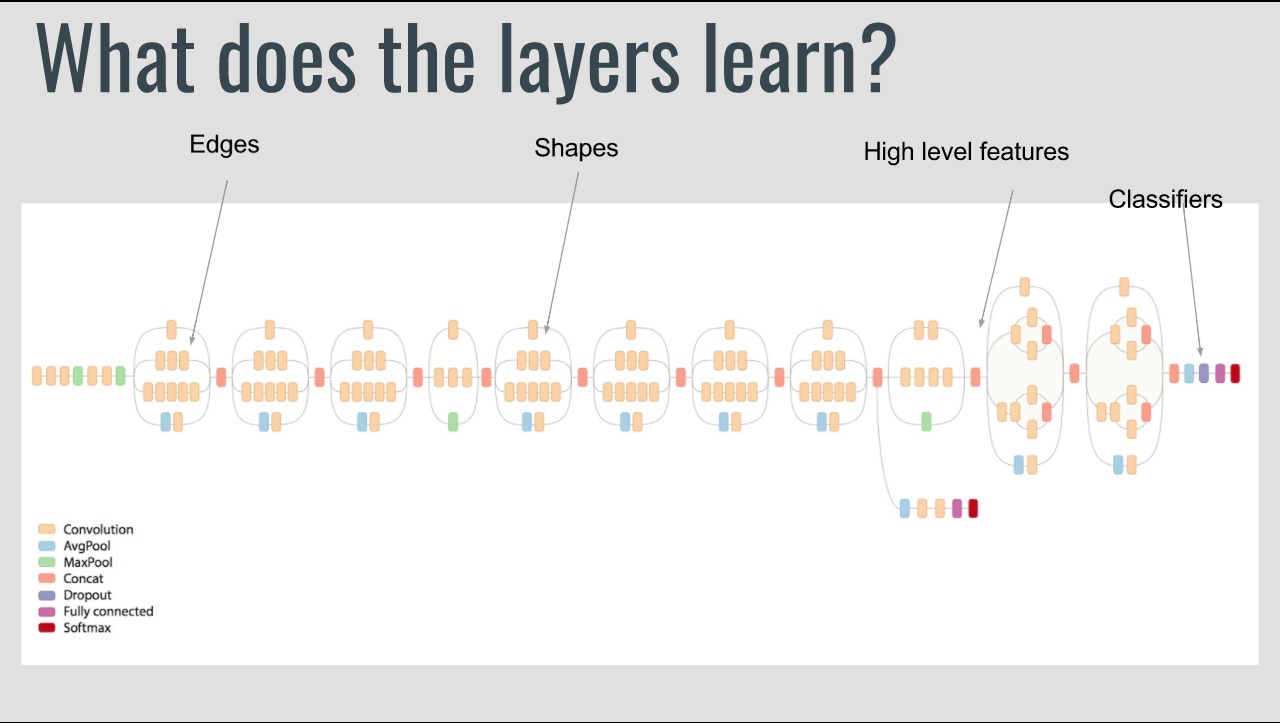
We are testing Inception, RESNET and Alexnet CNN architectures and will carry out training of the Neural Network on a high performance device (one time task). The initial case would be done using python with tensorflow, opencv, and numpy. The trained neural network architecture would be rebuilt in low-level, and the trained-weights would be borrowed and directly used.
Our key customers would be hobby gardeners, automatic greenhouse manufacturers/owners, and large-scale farmers..
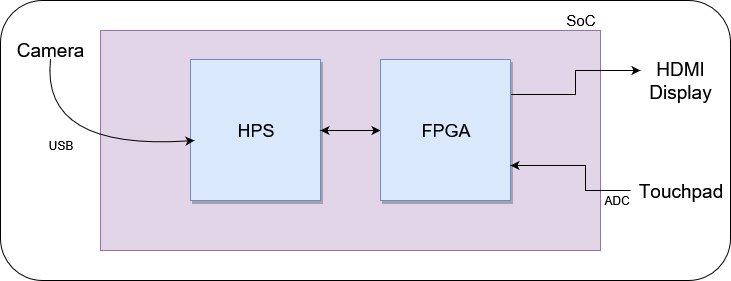
The overall system that would be built finally is as follows. The device would contain a single board computer and a FPGA. In the case of using the FPGA board, we will be using the on board SOC.
.png)
The hard processor system (HPS) acts as a controler of the whole system. It captures images through the camera connected through USB PHY interface and saves it onto the SPRAM, preprocesses it, and feeds it the FPGA. Also control functions from input to output (touch pad / display) to Neural Network (NNet) configuration is handled by the HPS.
.png)
The FPGA handles the core functions of the entire system: the NNet and input output pheripherals. Two controller modules are implemented on the FPGA:
The NNet controller handles all NNet functions. It reads input image from the SDRAM and results are sent back to SDRAM, and most importantly it handles configuration of NNet (initializing / updating weights).
The I/O controller handles the user friendly touch interface focussing on:
The I/O controller also interacts with the HPS and controls and executes functions of the system and fetches images from the SDRAM to be displayed on to the screen.
Since we have a convolutional neural network (CNN), several highly reusable blocks and a max controller are included. The CNN composes of convolution blocks, max pool blocks, densely connected linear blocks, non-linearity functions, and buffers. Each of these is built using block control units and highly reusable sub-units like Multiply-Add Units, Block RAM banks, Activation Functions (look-up tables), and Accumulate/Multiply Units. Activation functions using look-up tables is easier as changes require only replacing initiation parameters. Employing these highly reusable blocks for performing matrix operations makes it easier to adjust the layers of the NNet quickly.
eg :- nnet
.png)
An example block is displayed below:
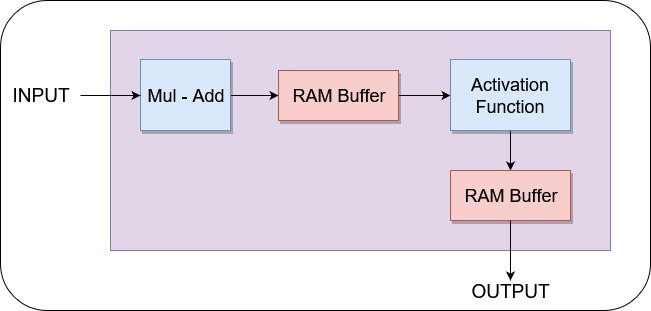
The process of building this system and integrating it with FPGA is described below.
Boost Performance
Our device would consist of a low performance single board computer. Inference of a Neural Network is a complex task that can be parallelized to boost performance. Hence we do this component on the FPGA. Also, offloading this work from the CPU allows the CPU to dedicate all resources to maintaining GUI and screen output.
Highly parallel computing nature of the FPGA allows real time interaction with user while consuming lower amount of power. Due to abundant hardware resources and powerful processing capacity ,many tasks such as matrix operations, activation can be completed in few clock cycles. In comparison even though conventional processing platforms such as CPUs and GPUs have higher clock speeds it takes very large number of clock cycles to perform above mentioned task while consuming large amount of power, thus making the FPGA the most suitable candidate for high performance mobile computing. Combined performance characteristics of FPGA enables implementing forward and backward algorithms in Convolutional neural networks. Hence using a FPGA allows this system to be used in real time mobile systems such as robots and UAVs.
In addition, we also hope to off load certain basic image processing onto the FPGA that would enable faster processing as well.
Scalability
Also, using FPGA means the ability to increase the complexity of the Neural Network used for the classification task. We can always increase the scale of our system. Further, as opposed to using dedicated chips, the FPGA gives us the ability to update the weighs of our neural network over time. This would be essential as changes are required when new diseases occur, or our performance is improved over time by applying new techniques and collecting larger datasets.
I/O Expansion
Inbuilt ADC, HDMI interface allows for a highly versatile user interface which makes the operation of the system extremely easier. And also creating custom controllers such as touch controller and display controller which are tailor made for our required application is very easy compared to conventional implementations where most of the time fully custom implementations are almost impossible and costly(Require complex circuits and an array of different controller ICs).
Automatic plant disease recognition has a wide range of applications in the modern agriculture context. From automated green houses to using drones in large farming fields, the technology of automatic plant disease recognition is a key component in merging the farming and agriculture industry with AI and IT.
Automatic plant disease recognition can be used by farmers to watch over their crops. It will become an essential component of drones used for surveying and taking care of huge fields of crops, whereas the home gardeners can use automatic plant disease recognition on mobile platforms as an expert solution to take care of his plants and trees. Whenever there is a lack in knowledge or resources, inability to identify or even something like automation and optimization, plant disease recognition using AI techniques is the key solution.
Our targeted user base extends through a vast variety of stakeholders.
One of the major issues of building a state of the art detection and recognition system is the issue of getting real time performance. FPGA allow us to accelerate the hardware of any system, thereby allowing us to implement the entire system on a real time basis. In addition, the ability to use Intel Altera FPGA cores gives us the ability to easily take our product to a state-of-the-art level in terms of optimizing some essential sub-processes.
Key Device Functions
The key functionality of the device is threefold and is listed out below.
Implementation
The first component of our device functionality is Anomaly Detection. This uses two camera feeds (NIR / RGB), extracting one band from each feed (the R band and NIR band) and combined there to form a large matrix containing all input data. Matrix operations that can be optimized with the FPGA are used for the computation of necessary indexes. Currently, we only have the NDVI index computed for our work.
These indexes allow us to gain a range of insight regarding the plant health, and presence of various diseases in a large open field. The advantage is that distant aerial images can be used to easily compute these indexes.
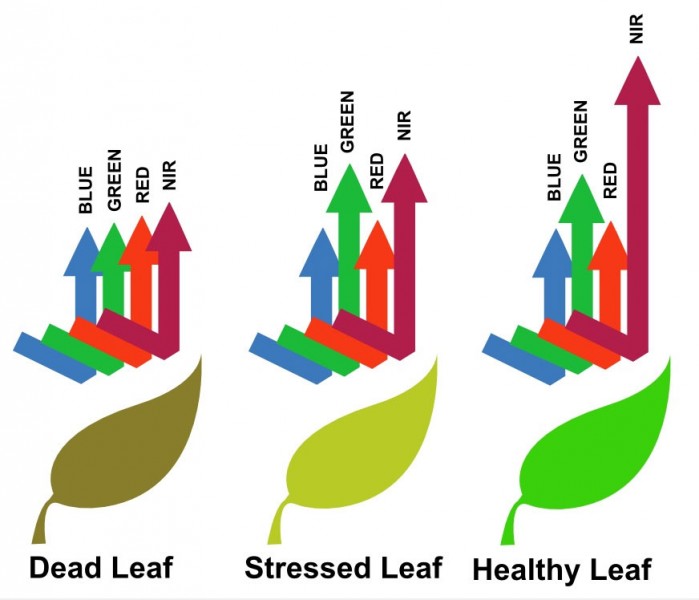
The next component in functionality is the exact image disease identification. Here an image obtained is first pre-processed to eliminate unwanted parts, and also ambient light based noise. Afterwards, this is fed into a neural network (convolutional neural network) pre-trained for unhealthy plant classification. We have trained our model on tomato plants and diseases for that taking into account seven different diseases and a healthy class as listed down below.
The device assigns a probability to each class. This is done through the Neural Network taking as input an image of a tomato plant leaf and outputting 8 probabilities to the eight classes. The problem is turned into a classification task and solved using the nerual network.
Running a complex neural network in real-time or close to real-time even is difficult due to computational complexity of the task. This problem is elevated when embedded devices need to run such algorithms. Hence, the FPGA based optimization in terms of speed is essential to procide this functionality. The entire inference of our pre-trained neural network is run using the FPGA.
The final component is the remote controllability built using a connection to a remote server and a web-app based interface. All data is uploaded to a remote server, and this allows the device to be accessed if it is connected to a drone or mounted onto a greenhouse roof.
The end user can access the device data using a web-app based interface as shown below. The plant disease, remedy and all other details are displayed. The example below is the basic interface.
The current web-app based interface is hosted here: https://plantcaredoc.com/webapp/.
Introduction
The performance of our device can be evaluated on two metrics; firstly, the overall accuracy in the system with regards to disease identifications, and secondly the efficiency of the system in terms of speed, energy consumption, and memory usage.
The project has two key components in terms of functionality, the anomaly detection and disease identification. The performance evaluations will be carried out for each component.
Anomaly Detection
With regards to the anomaly detection component, the output itself is an image, showing the NDVI index computed for each point in the image. The anomalous regions (that correspond to unhealthy plants) would  be highlighted. Hence, we calculate a direct accuracy: ratio of correct diseased regions highlighted to total regions highlighted by the device. In order to speed up the process, the image data representation (floating point) is compromised: basically, less decimal places would be recorded to improve speed. This results in a reduction of the accuracy. However, FPGA based operation allows a better speed, so we can use more longer representations as well. So, we compare the accuracy on a raw Raspberry Pi version as well as the version with our FPGA board (DE 10 Nano). Frames per second (FPS) processed are recorded. An average has been calculated after multiple trials.
be highlighted. Hence, we calculate a direct accuracy: ratio of correct diseased regions highlighted to total regions highlighted by the device. In order to speed up the process, the image data representation (floating point) is compromised: basically, less decimal places would be recorded to improve speed. This results in a reduction of the accuracy. However, FPGA based operation allows a better speed, so we can use more longer representations as well. So, we compare the accuracy on a raw Raspberry Pi version as well as the version with our FPGA board (DE 10 Nano). Frames per second (FPS) processed are recorded. An average has been calculated after multiple trials.
|
Accuracy |
Raspberry Pi version |
DE 10 Nano Version |
|
90 % |
0.5 FPS |
8 FPS |
|
85 % |
1 FPS |
12 FPS |
|
75 % |
1 FPS |
16 FPS |
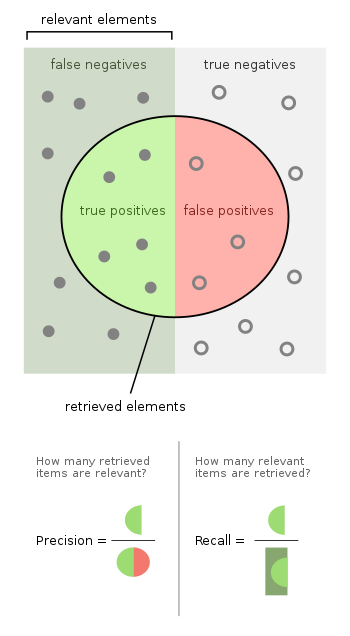 We should note that even on the case of a perfect NDVI index calculated, there can be errors in detection. Also, we noted that this direct accuracy does not show us the true positives our system failed to identify. Hence, we also calculated a precision and recall too which measures those component as well. This was only for the DE 10 Nano Version maintaining a 12 FPS rate.
We should note that even on the case of a perfect NDVI index calculated, there can be errors in detection. Also, we noted that this direct accuracy does not show us the true positives our system failed to identify. Hence, we also calculated a precision and recall too which measures those component as well. This was only for the DE 10 Nano Version maintaining a 12 FPS rate.
|
|
Precision |
Recall |
|
Trial 01 |
0.87 |
0.86 |
|
Trial 02 |
0.85 |
0.88 |
These are the current performance parameters of this component of the device. We hope to reach 90% on both of these metrics.
With regards to the power consumption and memory usage, we have been unable to make evaluations with regards to our device. The expected performance in these aspects needs to be within bounds to allow this to be deployed as an embedded device.
Disease Identification
This component uses a convolutional neural network. The task of image disease identification is treated as a classification problem, and the neural network is trained for this purpose using a large dataset of plant disease images. Here we simply calculate the accuracy of the system for our image dataset. The accuracy obtained is at 92% for well pre-processed data and ranges above 80% for pre-processed data from the wild.
Also, the FPGA based optimization allows us to run inference on an image at a speed of 1-2 FPS. We hope to improve it to a higher rate with more optimization. The current bottlenecks are in memory usage and data movement.
All images referred to in evaluations that use a FPS metric are resized to 244 X 244 size.
The basic hardware components required for this project are the FPGA camera and the HDMI touch display. The following block diagram shows how the components are interconnected.
The hard processor system (HPS) inside the FPGA acts as the controller of the whole system. It captures images through the camera connected through USB PHY interface and saves it onto the SPRAM.
Captured image is pre-processed in the HPS part and then fed in to a Convolutional neural network. Finally the output probabilities are displayed using the touch screen display which does the following two tasks.